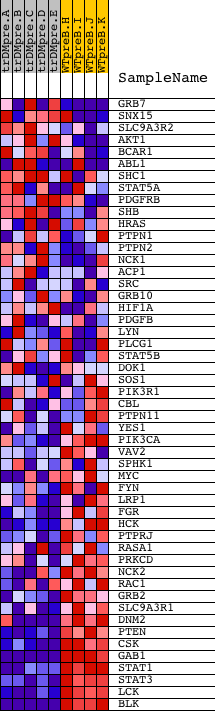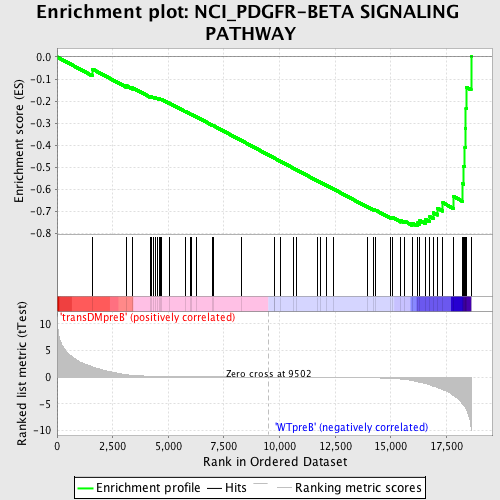
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | NCI_PDGFR-BETA SIGNALING PATHWAY |
| Enrichment Score (ES) | -0.76504046 |
| Normalized Enrichment Score (NES) | -1.6993846 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.025516812 |
| FWER p-Value | 0.087 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GRB7 | 20673 | 1581 | 1.934 | -0.0546 | No | ||
| 2 | SNX15 | 23993 3761 | 3104 | 0.460 | -0.1293 | No | ||
| 3 | SLC9A3R2 | 23089 1544 1514 | 3401 | 0.330 | -0.1400 | No | ||
| 4 | AKT1 | 8568 | 4184 | 0.164 | -0.1795 | No | ||
| 5 | BCAR1 | 18741 | 4251 | 0.158 | -0.1806 | No | ||
| 6 | ABL1 | 2693 4301 2794 | 4314 | 0.151 | -0.1816 | No | ||
| 7 | SHC1 | 9813 9812 5430 | 4421 | 0.139 | -0.1851 | No | ||
| 8 | STAT5A | 20664 | 4510 | 0.131 | -0.1877 | No | ||
| 9 | PDGFRB | 23539 | 4579 | 0.125 | -0.1894 | No | ||
| 10 | SHB | 10493 | 4664 | 0.118 | -0.1921 | No | ||
| 11 | HRAS | 4868 | 4707 | 0.115 | -0.1925 | No | ||
| 12 | PTPN1 | 5325 | 5029 | 0.095 | -0.2083 | No | ||
| 13 | PTPN2 | 23414 | 5764 | 0.062 | -0.2469 | No | ||
| 14 | NCK1 | 9447 5152 | 5996 | 0.056 | -0.2584 | No | ||
| 15 | ACP1 | 4317 | 6039 | 0.055 | -0.2598 | No | ||
| 16 | SRC | 5507 | 6271 | 0.049 | -0.2715 | No | ||
| 17 | GRB10 | 4799 | 6977 | 0.035 | -0.3089 | No | ||
| 18 | HIF1A | 4850 | 7022 | 0.034 | -0.3107 | No | ||
| 19 | PDGFB | 22199 2311 | 8287 | 0.016 | -0.3786 | No | ||
| 20 | LYN | 16281 | 9750 | -0.003 | -0.4573 | No | ||
| 21 | PLCG1 | 14753 | 9775 | -0.004 | -0.4585 | No | ||
| 22 | STAT5B | 20222 | 10037 | -0.007 | -0.4724 | No | ||
| 23 | DOK1 | 17104 1018 1177 | 10626 | -0.015 | -0.5039 | No | ||
| 24 | SOS1 | 5476 | 10751 | -0.016 | -0.5103 | No | ||
| 25 | PIK3R1 | 3170 | 11694 | -0.031 | -0.5606 | No | ||
| 26 | CBL | 19154 | 11843 | -0.033 | -0.5680 | No | ||
| 27 | PTPN11 | 5326 16391 9660 | 12087 | -0.038 | -0.5805 | No | ||
| 28 | YES1 | 5930 | 12441 | -0.046 | -0.5988 | No | ||
| 29 | PIK3CA | 9562 | 13936 | -0.107 | -0.6776 | No | ||
| 30 | VAV2 | 5848 2670 | 14239 | -0.130 | -0.6918 | No | ||
| 31 | SPHK1 | 5484 1266 1402 | 14328 | -0.138 | -0.6943 | No | ||
| 32 | MYC | 22465 9435 | 14983 | -0.233 | -0.7259 | No | ||
| 33 | FYN | 3375 3395 20052 | 15076 | -0.254 | -0.7268 | No | ||
| 34 | LRP1 | 9284 | 15456 | -0.363 | -0.7415 | No | ||
| 35 | FGR | 4723 | 15626 | -0.432 | -0.7438 | No | ||
| 36 | HCK | 14787 | 15955 | -0.616 | -0.7517 | Yes | ||
| 37 | PTPRJ | 9664 | 16204 | -0.867 | -0.7513 | Yes | ||
| 38 | RASA1 | 10174 | 16303 | -0.967 | -0.7413 | Yes | ||
| 39 | PRKCD | 21897 | 16559 | -1.188 | -0.7363 | Yes | ||
| 40 | NCK2 | 9448 | 16752 | -1.454 | -0.7237 | Yes | ||
| 41 | RAC1 | 16302 | 16919 | -1.693 | -0.7058 | Yes | ||
| 42 | GRB2 | 20149 | 17099 | -1.953 | -0.6846 | Yes | ||
| 43 | SLC9A3R1 | 11250 | 17339 | -2.365 | -0.6601 | Yes | ||
| 44 | DNM2 | 4635 3103 | 17822 | -3.423 | -0.6320 | Yes | ||
| 45 | PTEN | 5305 | 18232 | -5.045 | -0.5743 | Yes | ||
| 46 | CSK | 8805 | 18265 | -5.201 | -0.4938 | Yes | ||
| 47 | GAB1 | 18828 | 18330 | -5.503 | -0.4103 | Yes | ||
| 48 | STAT1 | 3936 5524 | 18359 | -5.675 | -0.3221 | Yes | ||
| 49 | STAT3 | 5525 9906 | 18377 | -5.818 | -0.2311 | Yes | ||
| 50 | LCK | 15746 | 18405 | -6.029 | -0.1373 | Yes | ||
| 51 | BLK | 3205 21791 | 18607 | -9.402 | 0.0005 | Yes |